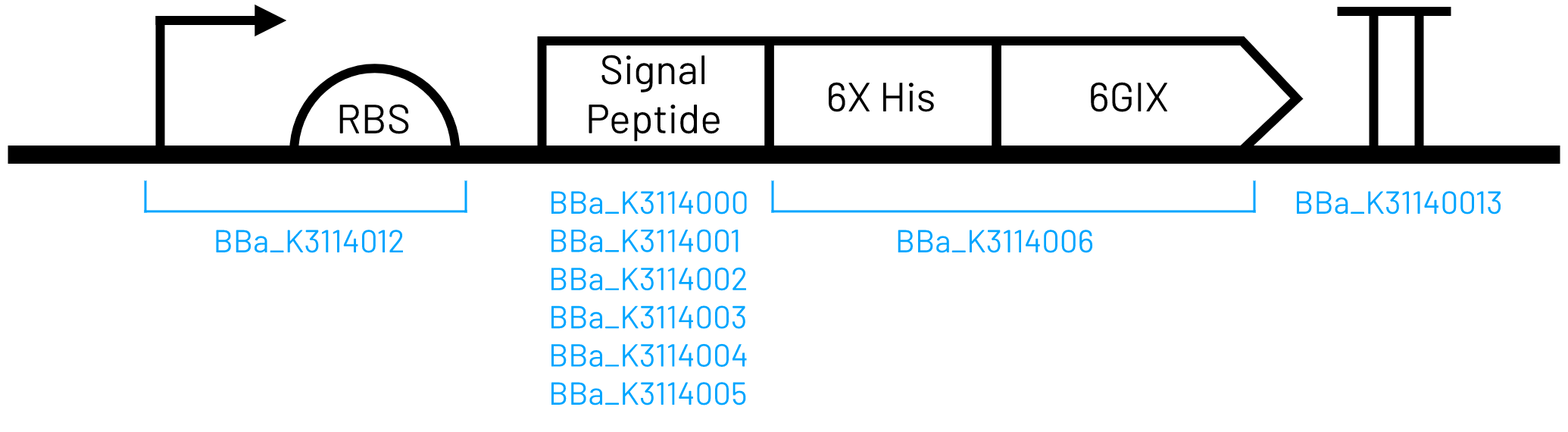
Part:BBa_K3114021
6xHis-tagged water-soluble chlorophyll binding protein (6GIX) circuit with TorA signal peptide
Usage and Biology
This part can be used for IPTG-inducible expression and secretion of the water-soluble chlorophyll binding protein 6GIX, which contains a 6xHis tag for purification. This circuit contains the TorA signal peptide.
iGEM Calgary successfully created seven inducible genetic circuits for high-level production of 6GIX using various parts from our collection.
- DsbA signal peptide 6GIX circuit (BBa_K3114016)
- MalE signal peptide 6GIX circuit (BBa_K3114017)
- OmpA signal peptide 6GIX circuit (BBa_K3114018)
- PhoA signal peptide 6GIX circuit (BBa_K3114019)
- YcbK signal peptide 6GIX circuit (BBa_K3114020)
- TorA signal peptide 6GIX circuit (BBa_K3114021)
- 6GIX circuit with no signal peptide (BBa_K3114022)
6GIX is a 180-amino acid water-soluble chlorophyll binding protein (WSCP) which is hypothesized to play a role as a transient chlorophyll shuttle or to be involved in anti-photobleaching responses in Lepidium virginicum (Takahashi et al., 2013). 6GIX is capable of binding chlorophyll a and b, but it has been shown to have higher affinity for chlorophyll b (Bednarczyk, Takahashi, Satoh, & Noy, 2015; Palm et al., 2018).
6GIX exists as a homotetramer that is capable of binding four chlorophyll molecules (Bednarczyk, Takahashi, Satoh, & Noy, 2015). Chlorophyll is a hydrophobic pigment and is therefore soluble only in organic solvents. This part can be used for aqueous phase capture of chlorophyll using emulsions.
The TorA signal peptide is a 39-amino acid sequence which targets fused proteins to the Tat secretion pathway in E. coli (Silvestro et al., 1989). In this pathway, proteins are fully or partially folded before secretion to the periplasm. The signal peptide is cleaved after residue 39 by signal peptidase after secretion (recognition sequence: Ala-X-Ala).
Design
When designing this circuit and the rest of our collection, we were interested in creating parts that could be used in Golden Gate assembly right out of the distribution kit without the need to first domesticate them in a Golden Gate entry vector. As such, the basic parts are not compatible with the iGEM Type IIS RFC[1000] assembly standard because we included the BsaI restriction site and MoClo standard fusion site in the part’s sequence.
This part was constructed using Golden Gate assembly and parts listed below. It is iGEM Type IIS RFC[1000] compatible because the BsaI sites are removed during assembly.
- T7 Promoter and strong RBS (BBa_K3114012)
- TorA signal peptide (BBa_K3114005)
- 6xHis-tagged 6GIX (BBa_K3114006)
- Double terminator (BBa_K3114013)
The circuit was designed for inducible, high-level expression and has been codon optimized for E. coli. A 6X Histidine affinity chromatography tag was added to the N-terminus of the 6GIX coding sequence for purification.
Characterization
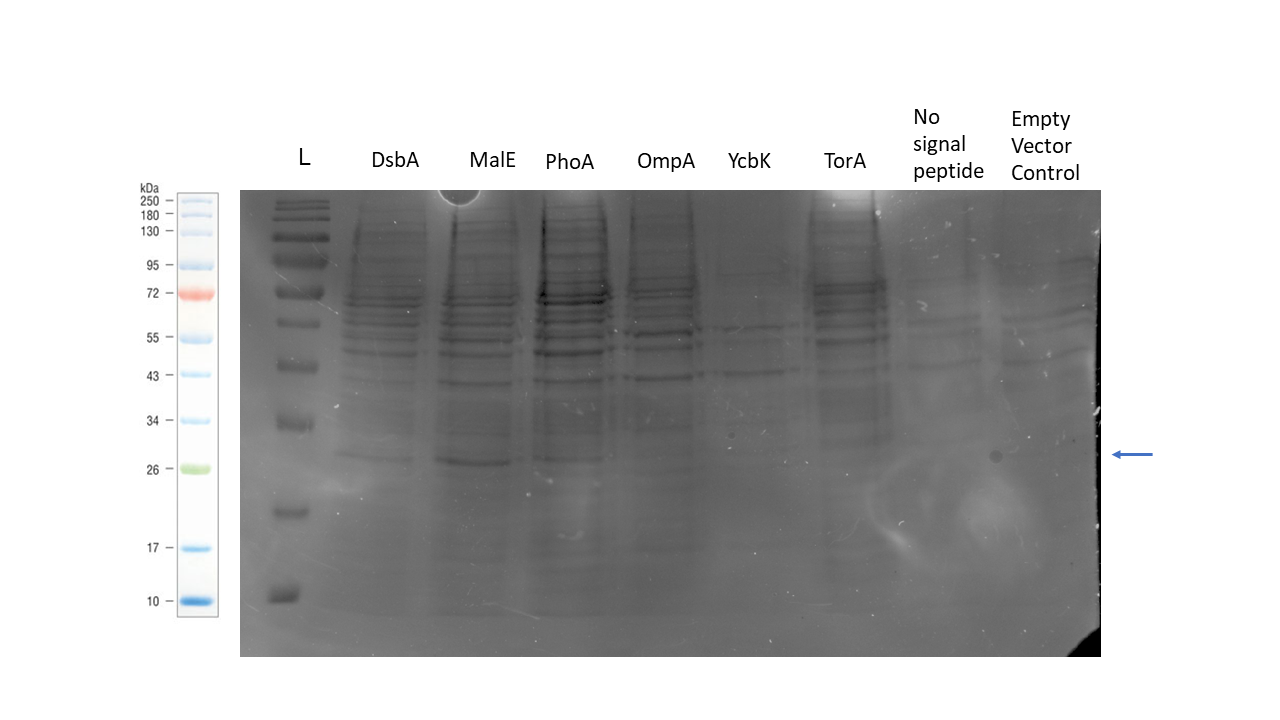
We were able to secrete 6GIX with the TorA signal peptide using this genetic construct. The SDS-PAGE gel below shows the protein recovered from the periplasm for each of our genetic constructs as per our periplasmic protein isolation protocol. The construct with the MalE signal peptide showed the strongest band of the correct size, but bands are also visible for the DsbA and PhoA signal peptide samples.
For experimental characterization of 6GIX’s function, please see (BBa_K3114006).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 761
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 286
Illegal AgeI site found at 229
Illegal AgeI site found at 545 - 1000COMPATIBLE WITH RFC[1000]
References
Bednarczyk, D., Takahashi, S., Satoh, H., & Noy, D. (2015). Assembly of water-soluble chlorophyll-binding proteins with native hydrophobic chlorophylls in water-in-oil emulsions. Biochimica et Biophysica Acta - Bioenergetics, 1847(3), 307–313. https://doi.org/10.1016/j.bbabio.2014.12.003
Palm, D. M., Agostini, A., Averesch, V., Girr, P., Werwie, M., Takahashi, S., … Paulsen, H. (2018). Chlorophyll a/b binding-specificity in water-soluble chlorophyll protein. Nature Plants, 4(11), 920–929. https://doi.org/10.1038/s41477-018-0273-z
Silvestro, A., Pommier, J., Pascal, M. C., & Giordano, G. (1989). The inducible trimethylamine N-oxide reductase of Escherichia coli K12: its localization and inducers. Biochimica et Biophysica Acta (BBA)/Protein Structure and Molecular, 999(2), 208–216. https://doi.org/10.1016/0167-4838(89)90220-3
Takahashi, S., Yanai, H., Oka-Takayama, Y., Zanma-Sohtome, A., Fujiyama, K., Uchida, A., … Satoh, H. (2013). Molecular cloning, characterization and analysis of the intracellular localization of a water-soluble chlorophyll-binding protein (WSCP) from Virginia pepperweed (Lepidium virginicum), a unique WSCP that preferentially binds chlorophyll b in vitro. Planta, 238(6), 1065–1080. https://doi.org/10.1007/s00425-013-1952-7
Weber, E., Engler, C., Gruetzner, R., Werner, S., & Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS ONE, 6(2). https://doi.org/10.1371/journal.pone.0016765
//chassis/prokaryote/ecoli
| None |